By Michael Mozdy
Western medicine has advanced from one-size-fits-all treatments like leaches for a fever to the frontier of personalized medicine, where each patient is treated differently, even those with the same diagnosis. This sounds great, but how exactly do we personalize medicine?
Genetic testing is one way to tell healthy patients if they have a risk for developing rare diseases or specific types of cancers. There has been much excitement in the past few decades about being able to “turn off” genes involved in disease or introduce helpful genetic therapies. However, little progress has been made in part because there are often many genes (not just one) that impact a disease state and affecting genes has unforeseen consequences. So, genetics has value in predicting a diagnosis (although since genes are also responsive to environmental factors, this is just a probability, not a certainty), and less value thus far towards treatment or monitoring.
In the meantime, the field of imaging science has introduced surprisingly powerful – and practical – ways to see the real-time biology of disease as it affects individuals differently.
 Since X-rays were first unveiled in 1896, noninvasive imaging like CT and MRI have produced better and better looking images, allowing physicians to diagnose injury and disease with more and more accuracy. At the same time, different types of imaging advanced. Instead of visualizing the structures in the body, molecular imaging like PET and SPECT visualize biological processes in the body.
Since X-rays were first unveiled in 1896, noninvasive imaging like CT and MRI have produced better and better looking images, allowing physicians to diagnose injury and disease with more and more accuracy. At the same time, different types of imaging advanced. Instead of visualizing the structures in the body, molecular imaging like PET and SPECT visualize biological processes in the body.
“We can measure metabolism, oxygen status, blood flow, cell growth, and many other processes,” states Dan Kadrmas, PhD, a faculty member of the Department of Radiology and Imaging Sciences and a researcher at the Utah Center for Advanced Imaging Research. By measuring these processes, physicians can see just how aggressive a tumor might be or how badly affected a region of diseased heart tissue may be. This type of information goes far beyond whether a person has cancer or heart disease – it tells us exactly where and how their disease is progressing, within that individual patient.
Kadrmas is intensely interested in using molecular imaging as a key tool in personalized medicine, and has mapped out a new technique for getting there.
The Need for Quicker, More Definitive Personalized Medicine
Kadrmas is an active man with a sharp mind who speaks fast but wants to make sure you’re following him. “Let’s take pancreatic cancer as an example of what functional imaging can do,” he begins. “It’s a nasty cancer with a 5-year survival of only around 8%. Patients with pancreatic cancer don’t have a lot of time to try different therapies or a conservative approach.”
The sooner pancreatic cancer is detected, the better, and the sooner treatments are started, the better. Gemcitabine-plus-Abraxane is a first-line chemotherapy combination for patients with pancreatic cancer.
“Some patients respond very well to GEM-plus-Abraxane, but it’s only effective in about 15% of patients,” Kadrmas explains. A number of factors contribute to this, including how the drug is delivered, how fibrous the tissue around the tumor is, and how much vasculature feeds the tumor region. Above all, though, researchers have discovered that tumors containing a large amount of a transmembrane protein called hENT1 are the ones that respond well to gemcitabine.
Patients are typically given gemcitabine along with other chemo drugs and then must wait precious time – up to 6 weeks – before having more tests to see if the drugs are having any effect on the tumor. “But we happen to have a molecular tracer that binds to hENT1,” exclaims Kadrmas, “and it may be able to tell us right up front whether gemcitabine will be effective.” In other words, you could do one quick scan before deciding which drugs to use – this is a much more personalized approach to medicine. “Better yet,” Kadrmas adds, “we could run other tests just days after treatment starts to directly measure how the tumor is or isn’t responding.”
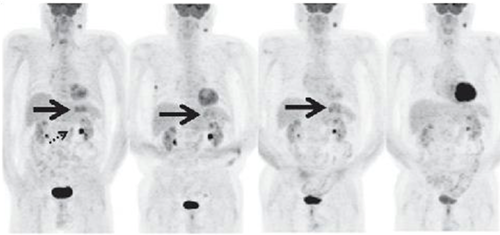
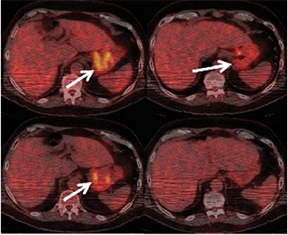
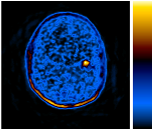
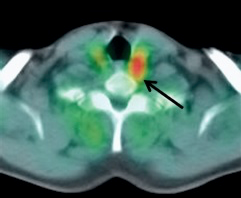
These PET scans (full body on left and cross-section on the right) show how a gastrointestinal tumor has responded to treatment in just one week. It would take several months before structural imaging such as CT scans could show that the tumor physically shrunk.
In short, molecular medicine can quickly impact treatment decisions and provide immediate knowledge of how treatments are working.
If personalized medicine was like the game Where in the World is Carmen Sandiego?, genetic testing could tell you if you’re likely to find her (but not where), while molecular imaging could tell you which street she’s walking down.
Tracing Molecular Usefulness
Kadrmas exudes an infectious enthusiasm for the potential of molecular medicine. The thrust of his argument is that one scan might be helpful, but the real power happens when multiple scans examine several biological processes simultaneously. He uses the example of how cancer spreads. “Cancerous tumors often grow fast and have a robust blood supply, but some of the most difficult ones to treat thrive in a low-oxygen environment,” he says, “and these are ones that often trigger metastases.”
With this information, physicians have several biological targets to create a complete picture of a patient’s health:
- Glucose metabolism, which happens voraciously in aggressively growing tumors, can be studied for tumor detection, tumor grading, and monitoring tumor response to therapies.
- Blood flow, which speeds up to feed some types of tumors, can show tumor growth and also be an indication that anti-angiogenic therapy may be successful or that drug delivery may or may not work.
- Low oxygen can mean the tumor is resistant to radiation therapy and some drugs, and it might be a metastatic trigger.
- Cellular proliferation, which helps “grade” tumors and could suggest that anti-proliferative therapy might help; it is also a way to monitor tumor response to any therapy.
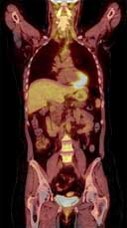
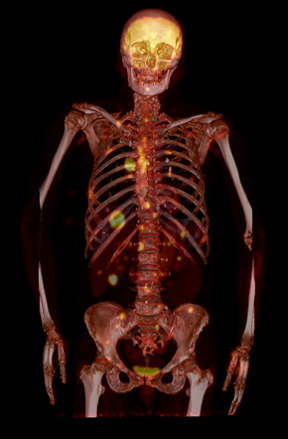
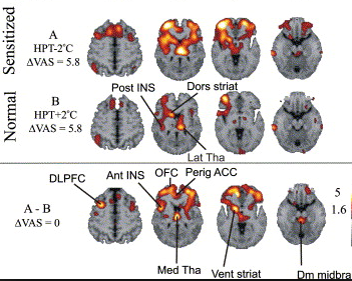
Types of scans and tracers: (top row, left to right) FDG for glucose metabolism, F18 bone scan for oxygen, 15O water for blood flow, (bottom row) FLT for cellular proliferation, 11C Methionine for amino acids.
The problem is that examining these processes in real time in a particular patient isn’t like doing a battery of blood tests. A single PET scan with one tracer takes 30-90 minutes, with 6 hours of no food or water beforehand. If a physician wants to measure more than one process to get a better understanding of a tumor, this means several days of scans, inconvenience, and expense. It’s not practical or easy, and effectively creates a hard limit on how much physicians use molecular imaging to diagnose and monitor treatment.
“If we could multiple PET images in one scan, that would be the holy grail,” declares Kadrmas. He foresees a paradigm shift that would launch molecular imaging as a practical, effective tool for personalized medicine.
Cost and inconvenience create a hard limit on how much physicians use molecular medicine. Multi-tracer studies can change that.
What Ultramarathons and Research Have in Common
Kadrmas once thought multi-tracer PET was just a pipe dream, but his work has made it a reality. He tells his story through the lens of an ultramarathon runner. It’s not just the fact that 100-mile trail races and research take equivalently long times in career miles. Kadrmas also points to the psychological ups and downs along the way. The Wasatch 100, which he ran in 2015, isn’t called “100 Miles of Heaven and Hell” for nothing.
 “About two thirds of the way into the race, I hit the wall,” he admits. “The grueling climb toward Catherine’s Pass had completely done me in. I sat there in the freezing darkness at 10,000 feet with my wife and friend massaging my legs and encouraging me to go on. It took 20 minutes to convince that I could continue putting one foot in front of the other and move on, but I finally did.”
“About two thirds of the way into the race, I hit the wall,” he admits. “The grueling climb toward Catherine’s Pass had completely done me in. I sat there in the freezing darkness at 10,000 feet with my wife and friend massaging my legs and encouraging me to go on. It took 20 minutes to convince that I could continue putting one foot in front of the other and move on, but I finally did.”
Likewise, Kadrmas had been climbing the mountain of making his multi-tracer PET algorithms practical and usable for 13 years, and by 2014, was fairly exhausted.
His journey involved wading into the hefty computations behind quantitative molecular imaging. When creating a quantitative movie of how a PET tracer moves throughout the body, a lot of difficult math needs to be applied. PET researchers analyze something called compartmental modeling, and they must “fit a curve” of the value they observe to the model.
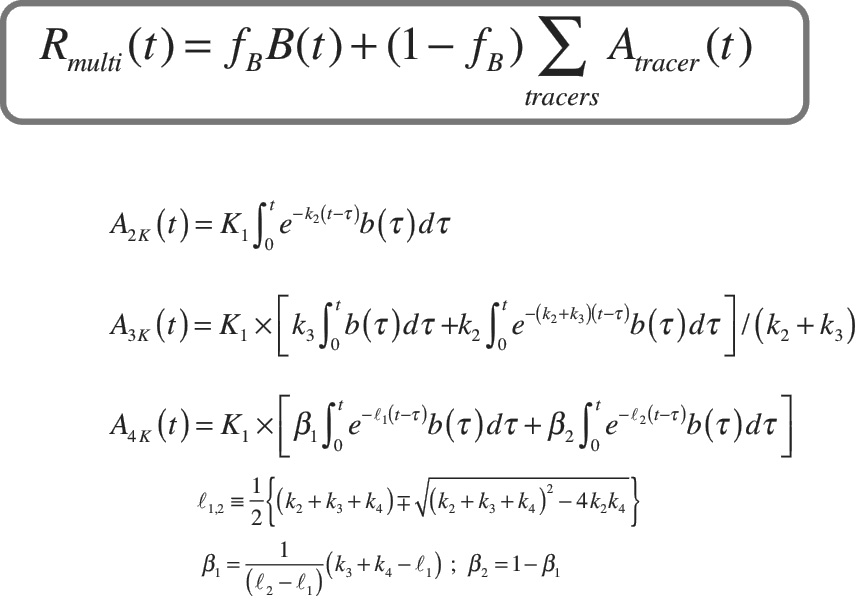 “Just fitting one curve in the past could take minutes to an hour of computer time, and you still got the wrong answer 80-90% of the time,” Kadrmas says. “We can now fit that curve in anywhere from five milliseconds to one second, and get the right answer 100% of the time.”
“Just fitting one curve in the past could take minutes to an hour of computer time, and you still got the wrong answer 80-90% of the time,” Kadrmas says. “We can now fit that curve in anywhere from five milliseconds to one second, and get the right answer 100% of the time.”
But this advancement refers to single-tracer fitting. “Multi-tracer modeling is 10-100x harder,” Kadrmas laments, “but we’ve gone from a processing time of 20 hours to 1 second and a success rate from under 50% to 100%.” The computational advancements suddenly made multi-tracer PET seem possible, and the next challenge was teasing out multiple signals from different radiotracers that were given around the same time.
Kadrmas has found that while not all of each signal could be evaluated as it would in single-tracer studies, 90% of the clinically relevant signal from each tracer could be analyzed. This small trade-off in thoroughness didn’t affect the overall clinical significance of the study, and the gains in time, cost, and patient comfort made it a no-brainer.
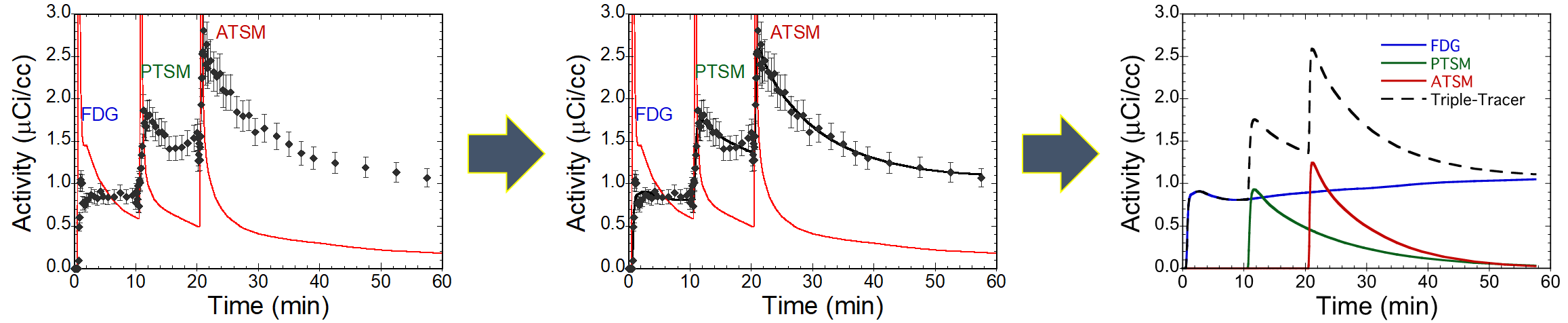
A visual representation showing Kadrmas’s work to “fit the curve” for multi-tracer PET studies.
“Fitting the multiple curves in a multi-tracer study is still a difficult process,” Kadrmas explains, “and must be re-designed for each combination of tracers that a physician might want to use.” In other words, if Kadrmas came up with a way to interpret the data from a lung cancer study using the three tracers FDG, 15O-water, and FLT, it would be applicable only in that study. If a physician wanted a study using FDG, 15O-water, and 18F-fluoromisonidazole, researchers would have to re-create and validate the approach for that particular study.
Kadrmas clearly saw the work that needs to be done in the coming decades to validate multi-tracer PET studies and attain widespread use. So, in 2014 he took the complicated algorithms developed over the last decade and pivoted to creating a platform to bring this technology into clinical practice.
He’s pointing towards the outfield fence, gunning for the home run: rather than proving how one multi-tracer study can be used in one specific scenario, he’s creating software that researchers around the world can use for many clinical trials of tracers chosen for specific cancers and diseases. It’s the tool that he hopes will open up the field for widespread adoption.
Kadrmas imagines a day when physicians will have a much bigger toolbox of FDA-approved multi-tracer PET studies they can use with their patients. These scans will provide extraordinarily precise data in real time, observing several biological processes in action.
This type of precision makes Kadrmas’s research and the entire field of molecular imaging so exciting for the future of personalized medicine. We anxiously look forward to the time when multi-tracer PET becomes a commonplace tool for physicians and patients.